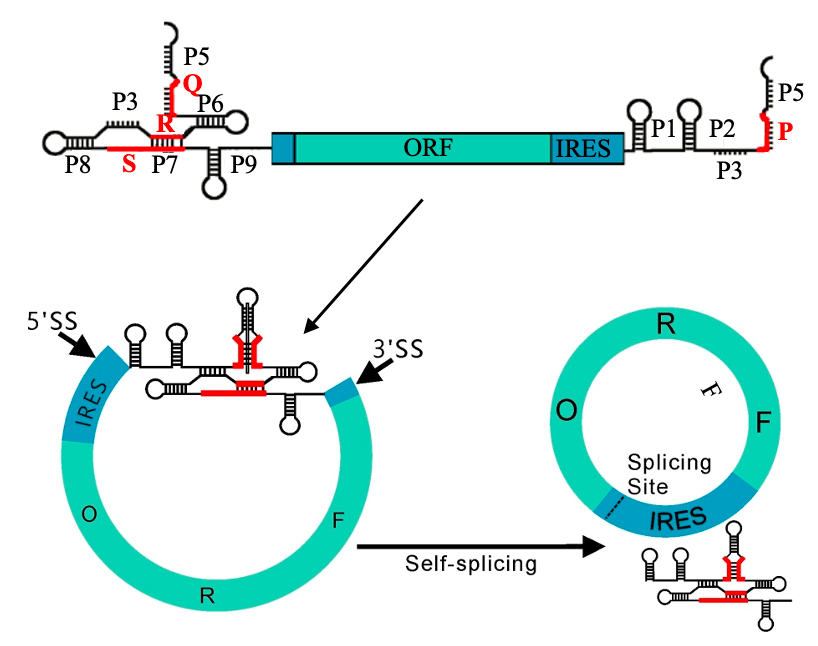
Clean PIE Circularization Frame

Since the outbreak of the COVID-19 epidemic, mRNA vaccines have undoubtedly become the most dazzling pearl in the crown of biomedicine due to its rapid, efficient and safe, wide-scale application. Thus mRNA technology has also been known to the public and has received attention from all levels of society. In the post-epidemic era, the application filed of mRNA has been continuously expanded, as the new indication requirements reflect many deficiencies of linear mRNA. Because of its closed-loop structure without requirement of capping and tailing, circular mRNA is more stable in vitro and in vivo, has longer expression duration, simpler preparation process and less cost. It is expected to become a substitute for linear mRNA and start the era of mRNA 2.0.
On June 21, 2022, SuZhou CureMed Biomedical Technology Co., Ltd. (hereinafter referred to as CureMed) published a paper on the preprint platform bioRxiv titled: Clean-PIE: a novel strategy for efficiently constructing precise circRNA with thoroughly minimized immunogenicity to direct potent and durable protein expression1.

The study disclosed an efficient, accurate, and low-immunogenic circular mRNA underlying circularization frame technology, called “Clean-PIE”, which achieves mass production scaling up and long-term continuous expression in vivo. Following ORNA and Laronde, CureMed has become a company of circular mRNA technology platform with underlying proprietary intellectual property rights and industrialization capabilities.
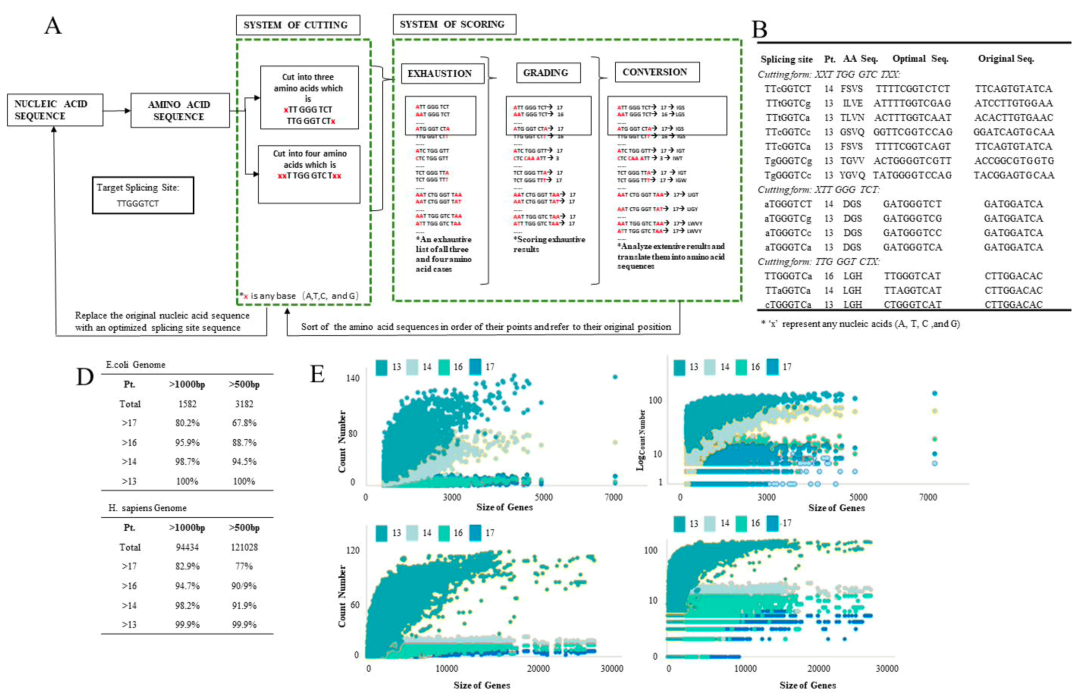
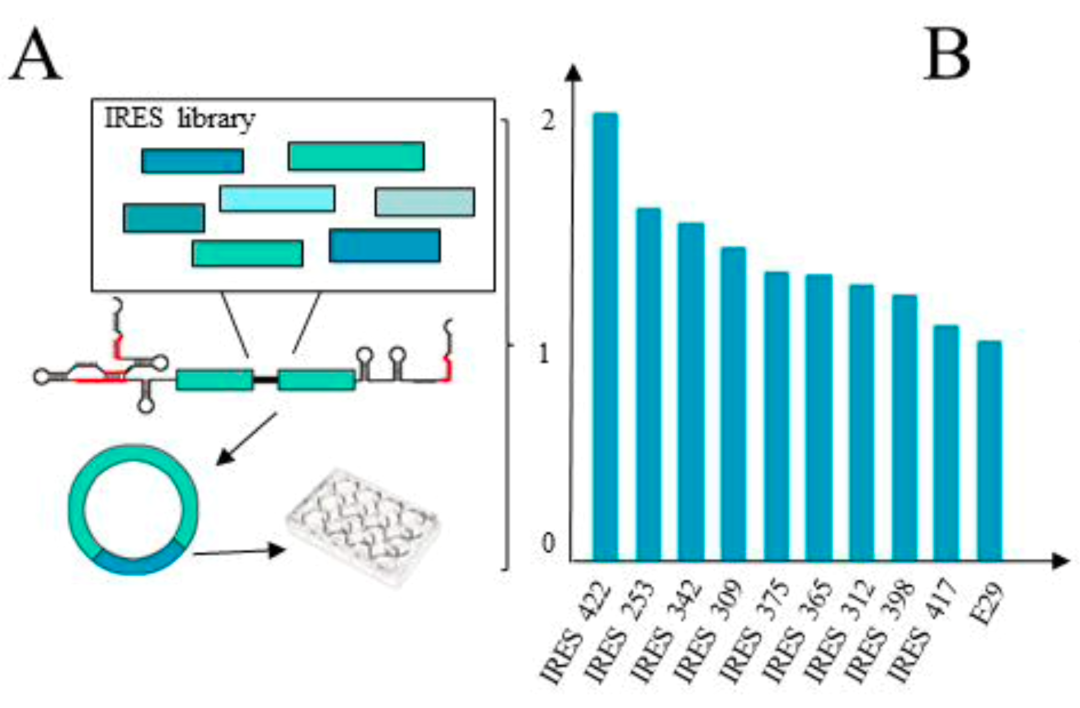
It is worth noting that the new Clean-PIE circularization strategy cleverly finds the optimal circularization point by screening the protein coding region or IRES region, and realizes the circularized connection of the protein coding region or IRES region without introducing exogenous sequences. Such strategy has high circularization effectivity (>90%) and accurate sequence; at the same time, the circular RNA obtained by this method is less immunogenic but of better expression efficiency and duration than that by ORNA. The study also establishes an automated splice point prediction and screening system for efficient computation of different sequences. In addition, it is worth mentioning that a high-efficiency IRES element screening system is established as well. After screening more than 600 IRES sequences, more than 20 new IRES sequences are obtained that are better than CVB3.
Circular RNA (circRNA) is a covalently closed circular RNA produced by the process of back-splicing in eukaryotes. In 1976, Sanger first discovered single-stranded and covalently closed circular RNA molecules in viroids, but it has long been considered the product of incorrect splicing of intracellular mRNA with no actual function. From the late 1990s to the early 20th century, studies found that many genes produced circRNAs. Since 2010, the development of RNA-seq technology has let off circRNA researches. In 2017, several research groups successively reported that circRNAs could be translated in a cap-independent manner in eukaryotes.
In the process when circRNA moves towards clinical application, a number of key problems are in front of us to be broken through:
1. How to achieve efficient and precise circularization of circRNA?
2. How to avoid the immunogenicity that comes with the introduction of exogenous sequences?
3. How to achieve efficient tissue-specific protein expression by circRNA?
4. How to obtain high-purity circRNA and avoid the interference of impurities such as precursor and nicked RNA?
5. How to achieve stable scale-up for mass production process?
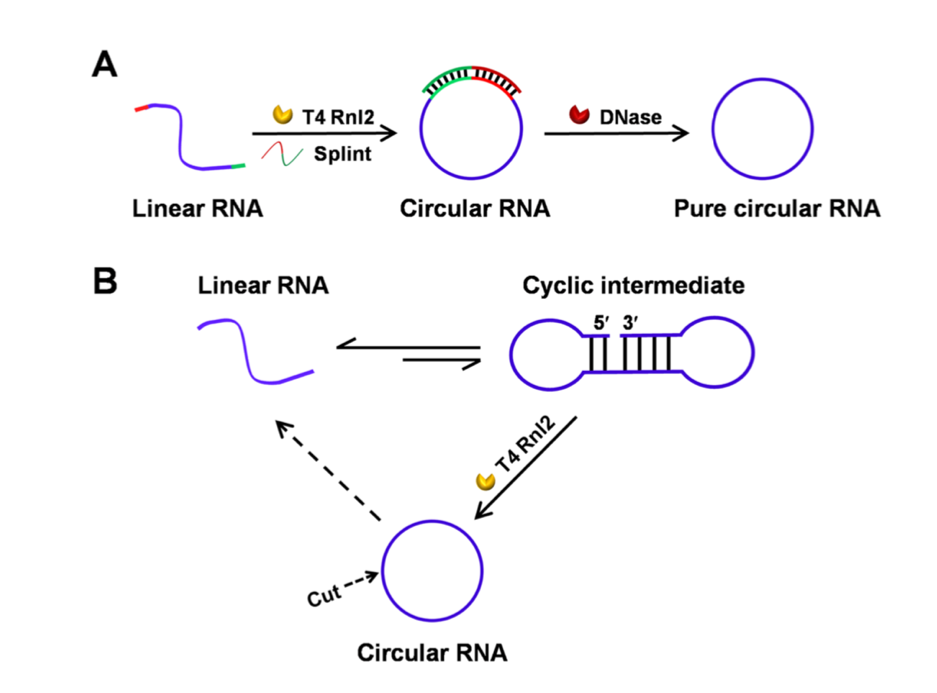
At present, there are three main strategies for in vitro RNA circularization: chemical method, ligase method (T4 RNA ligases), and ribozyme method (group I, group II self-splicing introns). The latter two methods are most commonly used.
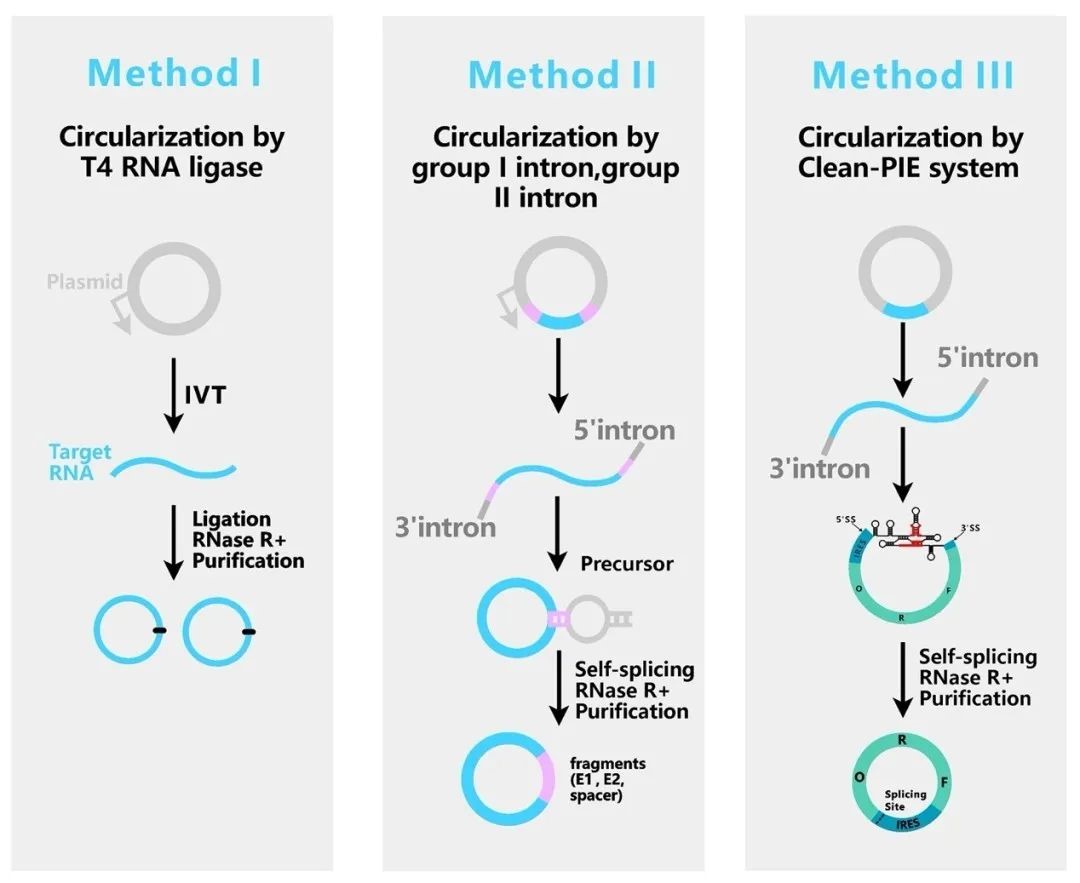
Method I: T4 RNA ligation
Method II: group I, group II intron self-splicing
Method III: Clean-PIE system, group I intron self-splicing
1. Group I intron self-splicing
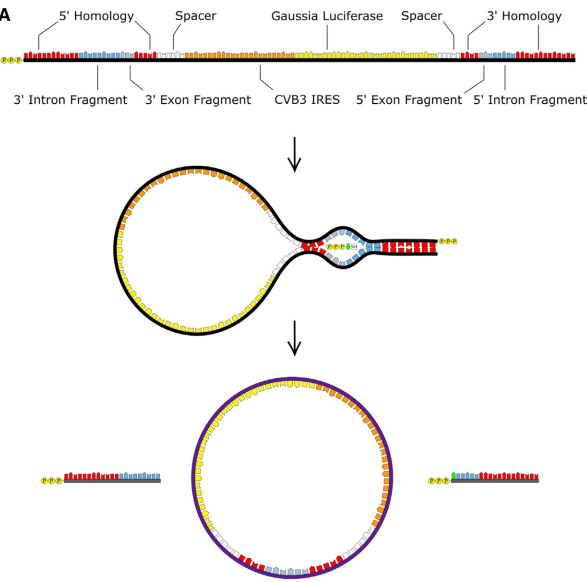
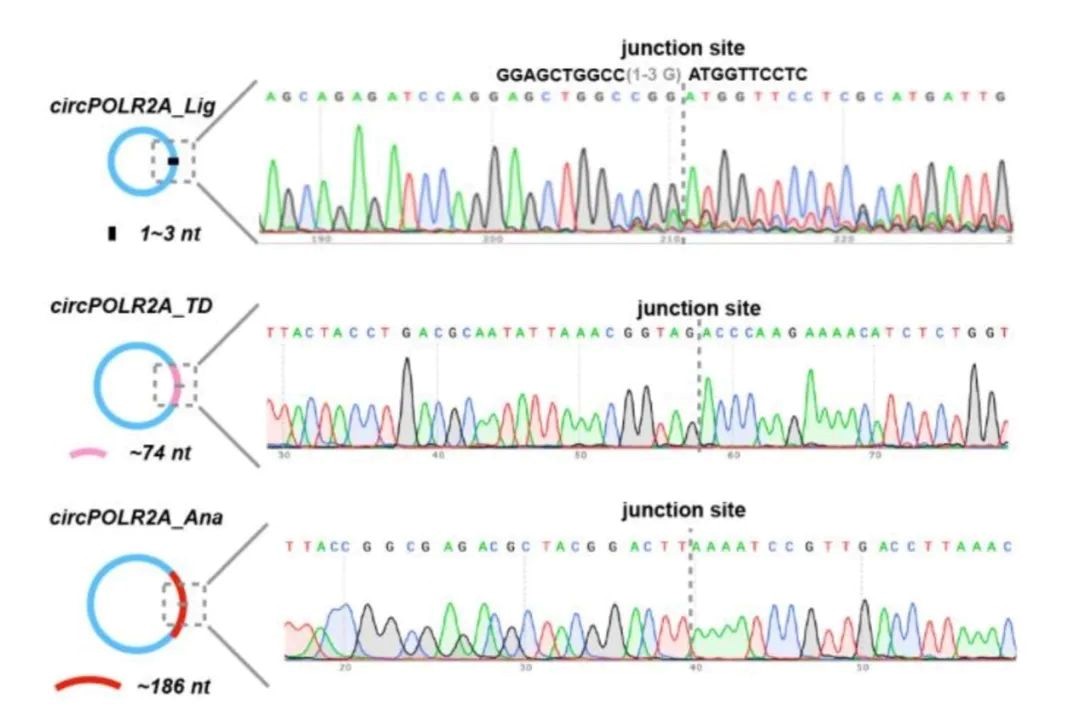
The research group of Daniel. G. Anderson2 in MIT achieved the circularization of long coding RNA in vitro by engineering transformation of Anabaena group I intron. In the presence of magnesium ions and GTP, two transesterification reactions occurred to form circular RNA. By optimizing the classic Anabaena PIE, adding homology arms and spacers, the problem of circularization for long coding RNA sequence was solved with good effectivity. Besides, the stability and efficiency of circRNA expression in cell protein were enhanced by HPLC purification strategy and IRES sequence screening. This circularization strategy requires an additional 186nt base sequence to be introduced into the final circular RNA product due to the addition of a spacer sequence and a longer exon sequence.
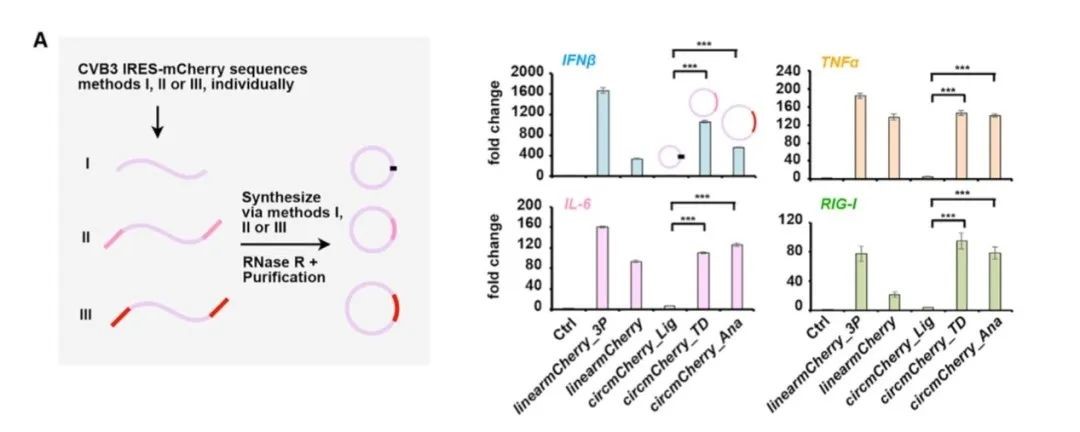
In March 2022, Professor Wei Wensheng of Peking University successfully prepared a COVID-19 RBD vaccine using the ORNA circularization frame, and observed excellent protection ability in rhesus monkeys against SARS-CoV-23.
In early 2022, Professor Chen Lingling published an article in Molecular Cell4 pointing out that back-self-splicing by Anabaena PIE or Thymidylate synthase (TD) PIE could cause significantly higher immunogenicity than the circRNA generated by the T4 ligase method, and proved that the immunogenicity was caused by additional sequences introduced by Ana PIE or td PIE during the circularization process. As shown below:
2. Group II intron self-splicing
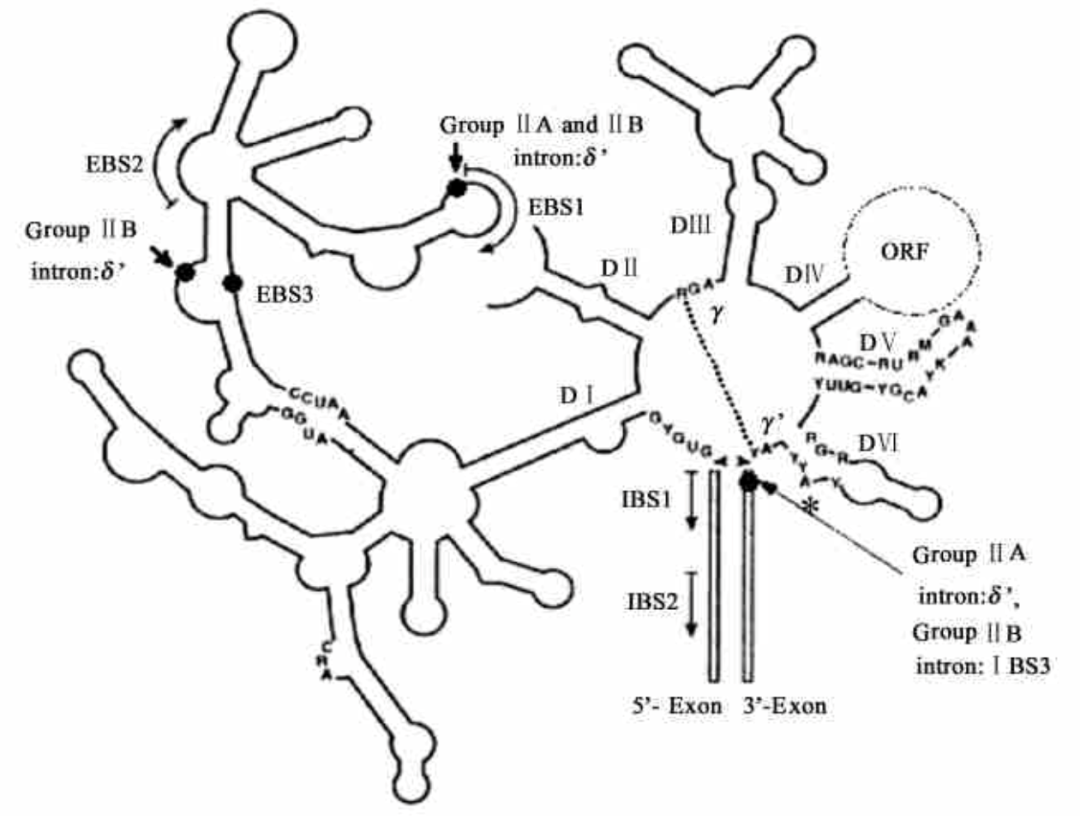
Group II introns are found in genomes of protists, fungus, and bacterias. Group II introns can be self-spliced from the precursor RNA via two consecutive transesterification reactions in vivo, and connect the flanking exons. Many group II intron splicing reactions are assisted by proteins. Meanwhile, group II introns have also been shown to undergo autocatalytic splicing reactions in vitro.
As shown in the figure below, the RNA target can be altered by the introduction of short IBS1 and IBS3 exogenous sequences by the autocatalytic splicing reaction of Bacillus tetanus group II intron, or by controlling the EBS sequences that pair with exons during intron insertion. Such modified group II introns are called “targetrons”. The splicing system of tetanus group II intron and yeast group II intron designed in this way can realize artificial in vitro preparation of circRNA without exogenous sequence residues5. As shown in the figure below, the circularization effectivity needs to be improved, and the nicked RNA contained in the circularized band needs to be further characterized.
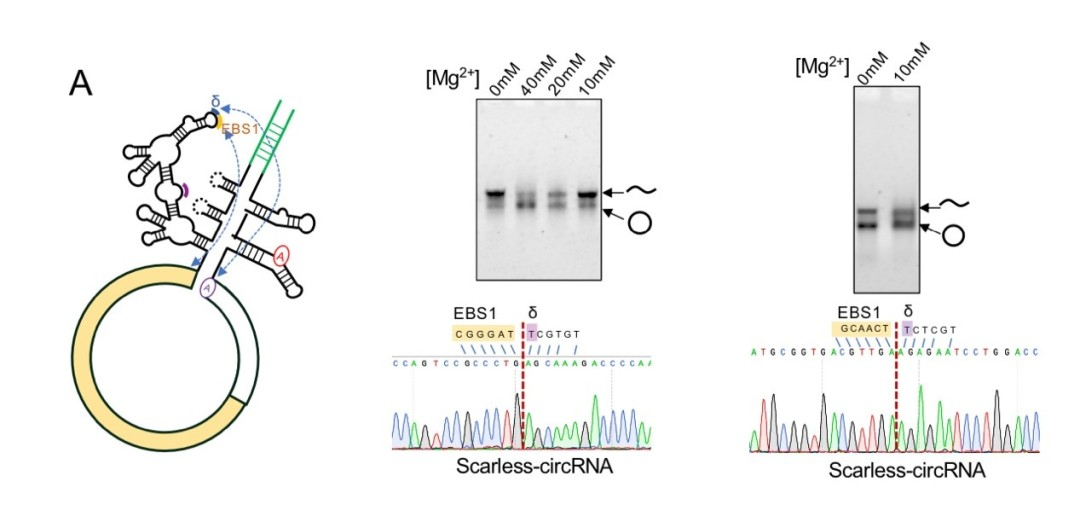
3. T4 RNA ligase
The in vitro circularization method of ligation of T4 DNA/RNA ligase is free of the immunogenicity caused by the introduction of exogenous sequences as above. However, the relatively low ligation efficiency of T4 ligase and the circularization method that requires the aid of DNA splint sequences make it difficult to achieve industrial production; this method also develops the circularization of RNA with the aid of endogenous splint strands3,6. As shown below:
Second, T7 RNA polymerase randomly introduces 1-3 additional guanine(G) 1 at the 3' end during transcription, so that the sequence obtained is inaccurate when the method of T4 RNA ligase is applied to the circularization of RNA transcribed in vitro by IVT;
Third, T4 RNA ligase catalyzes the formation of phosphodiester bonds between 5'-phosphate groups and 3'-hydroxyl ends among and within single-stranded RNA. After in vitro transcription, the 5' ends of the RNA strands need to be dephosphorylated in advance, which is cumbersome and inefficient. As shown below:
4. Clean-PIE group I intron self-splicing system
The immunogenicity caused by introduction of 186nt exogenous sequence by ORNA group I intron Ana PIE circularization strategy, the relatively low in vitro splicing efficiency of tetanus bacillus and yeast group II intron, the inaccuracy and cumbersome operation of T4 RNA ligase circularization method are all barriers that restrict the clinical application of circular RNAs.
On June 21, 2022, CureMed published a paper on the preprint platform bioRxiv, where a novel Clean-PIE circRNA circularization strategy was disclosed. The study cleverly found the optimal circularization point by screening the protein coding region or IRES region, and realized the circularized connection without introducing exogenous sequences, through the connection of protein coding region or IRES region .
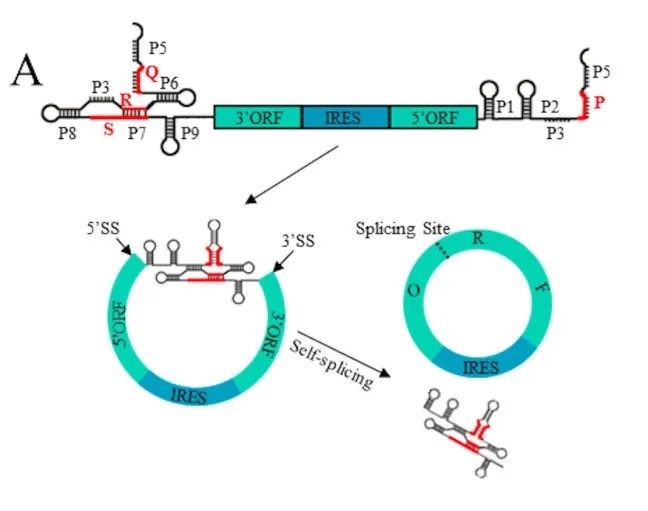
At the same time, it established an automatic screening and optimization system for splicing sites. The optimal circularization site was found out by efficient calculations for different sequences. By evaluating all genes larger than 500bp in the E. coli or human genome, it was found that the optimized system can obtain the circularization sites with more than 13 scores (that is, only one base away from the optimal looping sequence) by screening from 99.9% of the H. sapiens genes and 100% of the E. coli genes. This proves that the circularization strategy is universal.
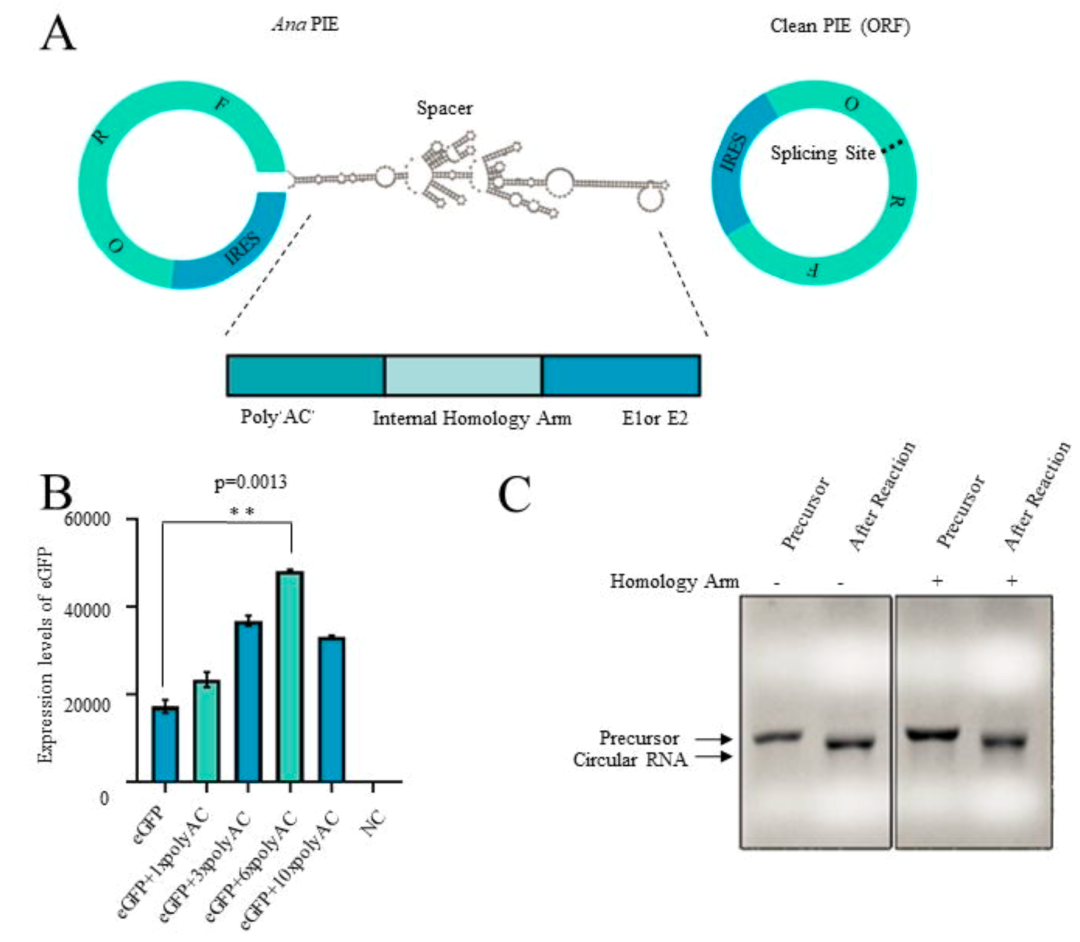
At the same time, this study optimized the sequence composition of circular RNA, and enhanced the expression of circular RNA by adding polyAC sequence.
The advantages of the Clean-PIE system are that no exogenous sequence is introduced, and the circularized sequence is accurate. The research data in this article show that compared with the ORNA circularization strategy, the intracellular expressions of IFN-β, RIG-1, IL-6 and MDA-5 in cells transfected with Clean-PIE circular RNA are lower, and the protein expression at the cellular level in vitro is also significantly better than ORNA.
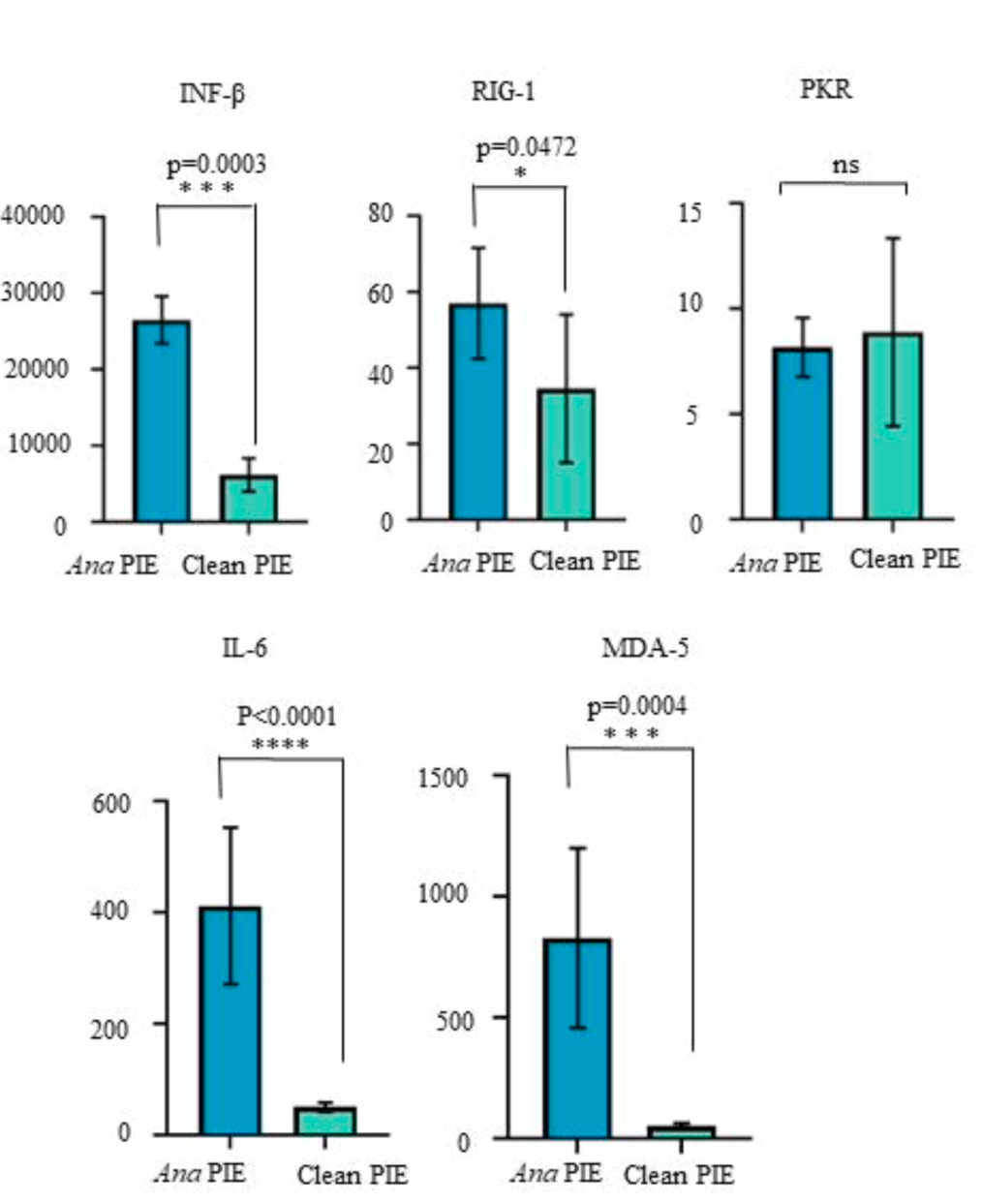
After purification by HPLC-SEC, both Ana PIE and Clean-PIE can achieve sustained protein expression in animals for up to 20 days. The length of time expressed in vivo is much better than that of linear mRNA.

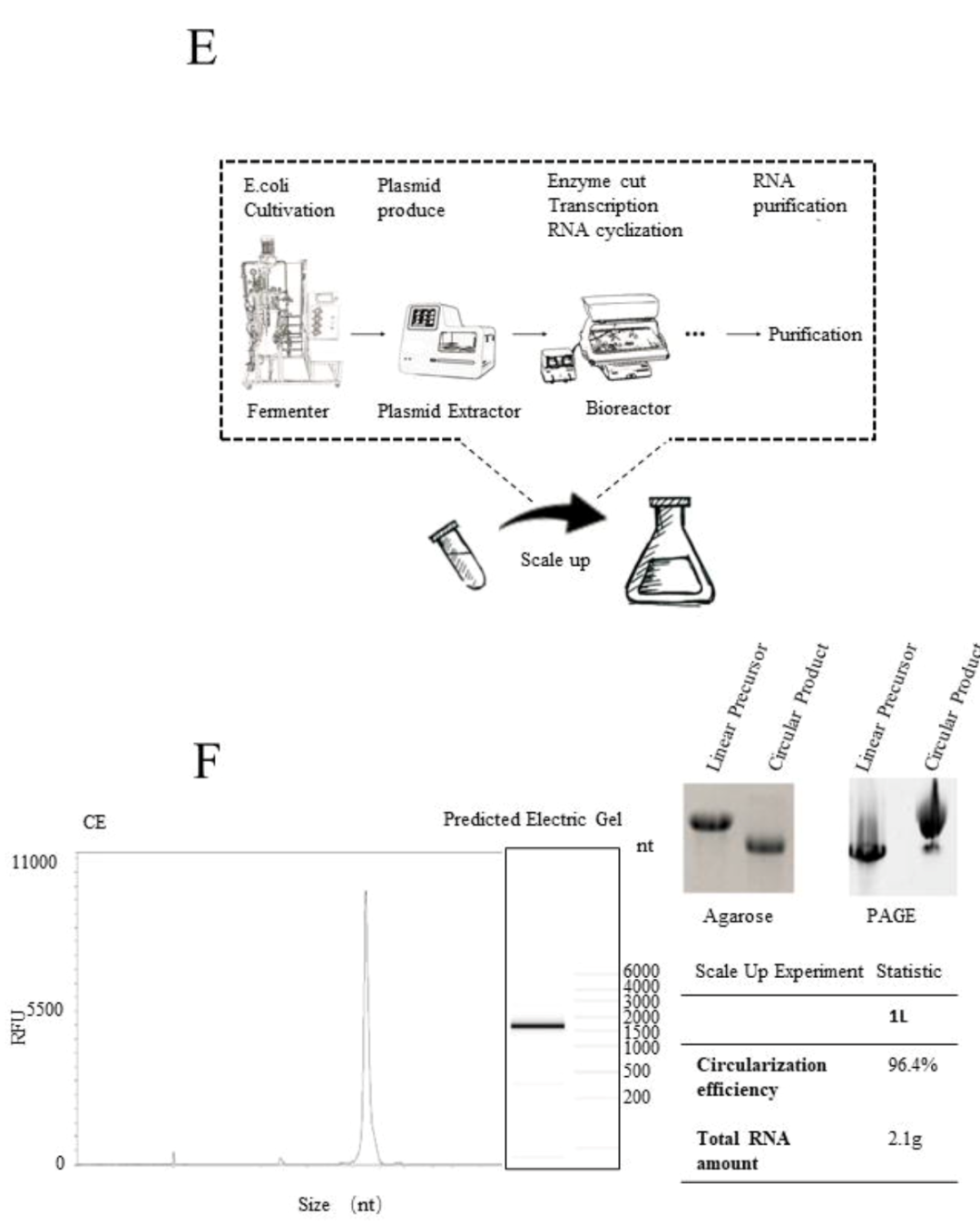
In addition, the study further carried out a scale-up experiment in a 1L reaction system. The capillary electrophoresis results showed that the circularization effectivity was as high as 96.4%, and the agarose gel electrophoresis results also proved the high effectivity. In order to further verify the production of nicked RNA in the circularization process, and evaluate the impurity residues of precursor and nicked RNA in the final product after purification, the purified circRNA was detected by Urea PAGE. Results proved that the circular RNA accounted for over 90% in the final product. That is to say, the Clean-PIE system is suitable for industrial mass production, and can ensure high circularization effectivity and high purity of final products. It lays the foundation for further clinical application.
In addition, in order to achieve high-efficiency tissue-specific protein expression of circRNAs, this study established an efficient IRES screening platform. More than 600 IRES sequences have been screened, of which more than 20 IRES elements have significantly higher expression levels than CVB3, and the IRES tissue-specific expression data have been primarily acquired.
Since November 2021, CureMed has successively published articles on cytokine intratumoral injection7, circular mRNA-based PROTAC technology8, new LNP nucleic acid delivery technology platform9, etc., demonstrating the completeness and maturity of LNP delivery system and circular mRNA platform technology, which are being rapidly applied to practical projects.
It is reported that the IIT research on circular RNA intratumoral injection project of FIRST IN CLASS is about to be advanced, and it is expected to take the lead in realizing the world's first circular RNA application in human.
The COVID-19 epidemic has accelerated the development of mRNA vaccines. As a platform technology, mRNA application is not limited to vaccines, but will be wider in tumor immunity, protein replacement, gene therapy and cell therapy. As the version 2.0 of mRNA technology, circular mRNA technology has attracted wide attention and aroused a trend of researches because it requires no capping, tailing or nucleotide modification, while performs more stably in vitro and in vivo with higher and longer protein expression.
Internationally, ORNA Therapeutics and Laronde are representative engagers in this field, especially when the latter has recently gained the B round finance of 440 million dollars from Flagship, which successfully incubated Moderna. ORNA, moreover, released its latest developments and the further in-depth arrangements in developing underlying patent technologies of IRES, LNP, etc. in the annual conference of ASGCT.
At home, researches are represented by the Clean-PIE group I intron self-splicing system of CureMed, the group II intron self-splicing system of CirCode, the Ana PIE of Therorna, and the T4 RNA ligase circularization method of Geneseed. Comparatively speaking, there is an R&D layout in quite distinct directions.
It can be expected that in the near future, there will be more and more circular mRNA research and development pipelines to meet the huge clinical demands and bring hope for patients to be cured.
[References]
1. Zonghao Qiu, Y. Z. et al. Clean-PIE: a novel strategy for efficiently constructing precise circRNA with thoroughly minimized immunogenicity to direct potent and durable protein expression..
2. Wesselhoeft, R. A., Kowalski, P. S. & Anderson, D. G. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat. Commun. 9, 2629 (2018).
3. Qu, L. et al. Circular RNA vaccines against SARS-CoV-2 and emerging variants. Cell. 185, 1728-1744 (2022).
4. Liu, C. X. et al. RNA circles with minimized immunogenicity as potent PKR inhibitors. Mol. Cell. 82, 420-434 (2022).
5. Chuyun Chen, H. W. Y. Y. A flexible, efficient, and scalable platform to produce circular RNAs as new therapeutics..
6. 梁兴国,陈辉,安然,程凯. 一种制备环状RNA的方法..
7. Jiali Yang, J. S., Jiafeng Zhu, Y. D., Yiling Tan, L. W., Qiangbo Hou, Y. Z. Z. S. & Chijian Zuo. Intratumoral Delivered Novel Circular mRNA Encoding Cytokines for Immune Modulation and Cancer Therapy..
8. Jiali Yang, J. S., Jiafeng Zhu, Y. D., Yiling Tan, L. W., Qiangbo Hou, Y. Z. Z. S. & Chijian Zuo. Circular mRNA encoded PROTAC (RiboPROTAC) as a new platform for the degradation of intracellular therapeutic targets..
9. Ke Huang, N. L. Y. L., Yuping Liu, Q. H. S. G., Ke Wei, C. D. C. Z. & Zhenhua Sun. Delivery of Circular mRNA via Degradable Lipid Nanoparticles against SARS-CoV-2 Delta Variant..
